Publications
2024
Automated construction of cognitive maps with predictive coding
2024, Nature Machine Intelligence

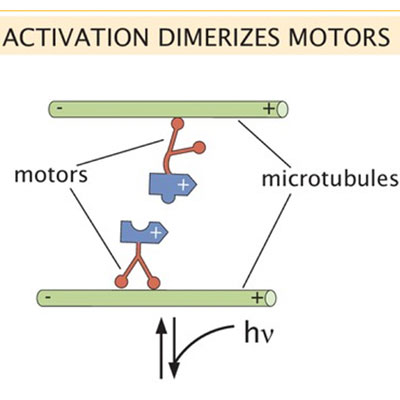
Active Healing of Microtubule-Motor Networks
2024, arXiv preprint
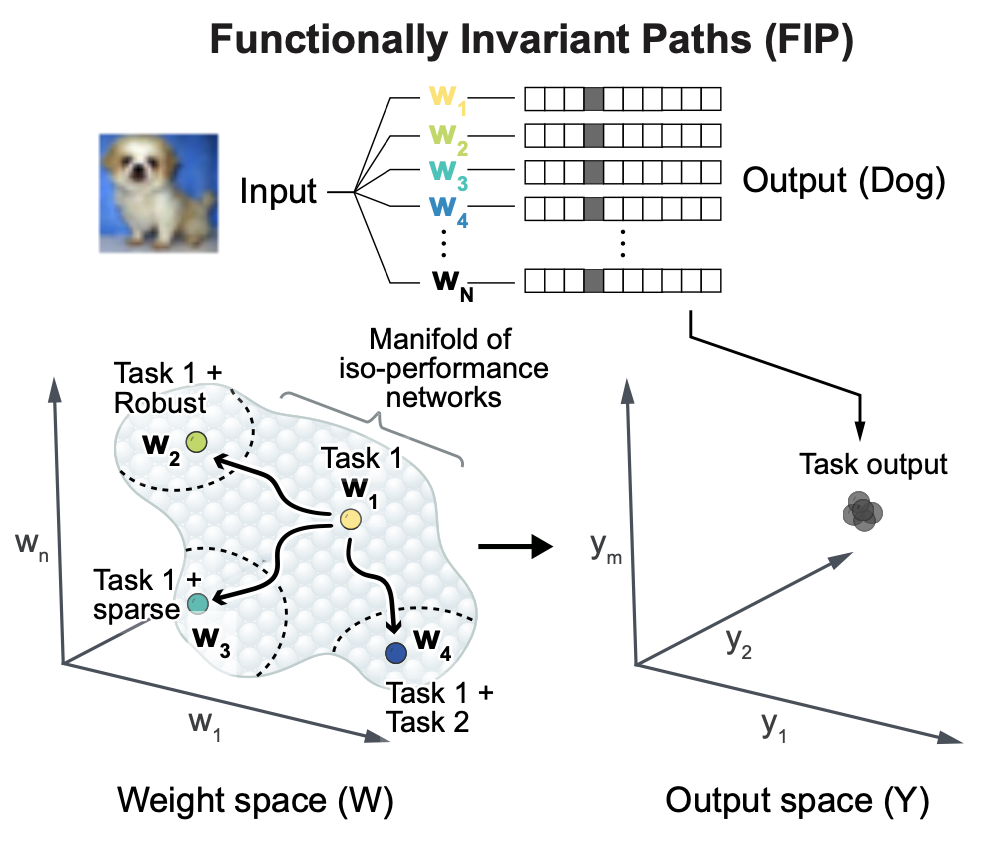
Engineering flexible machine learning systems by traversing functionally invariant paths in weight space
2024, Nature Machine Intelligence (Accepted)
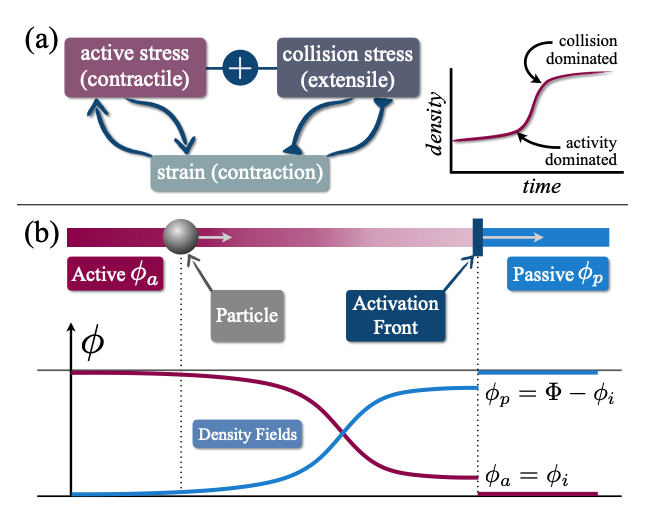
Theoretical Limits of Energy Extraction in Active Fluids
2024, Physical Review Research
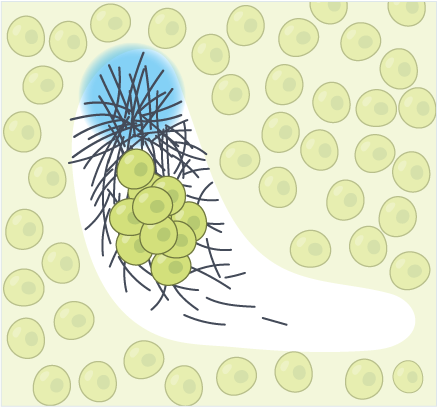
Force Propagation in Active Cytoskeletal Networks
2024, arXiv preprint
2023
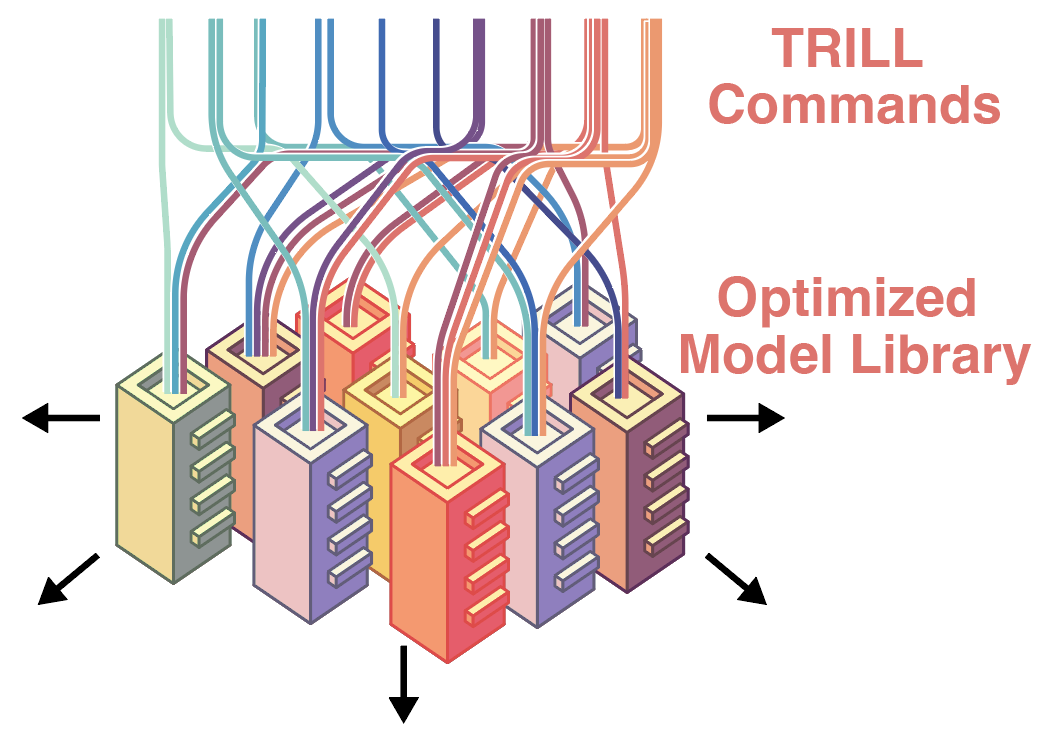
TRILL: Orchestrating Modular Deep-Learning Workflows for Democratized, Scalable Protein Analysis and Engineering
2023, bioRxiv preprint
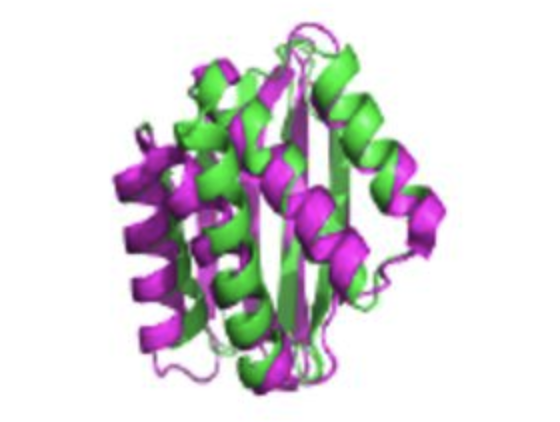
Unexplored regions of the protein sequence-structure map revealed at scale by a library of foldtuned language models
2023, bioRxiv preprint

What's the Magic Word? A Control Theory of LLM Prompting
2023, arXiv preprint
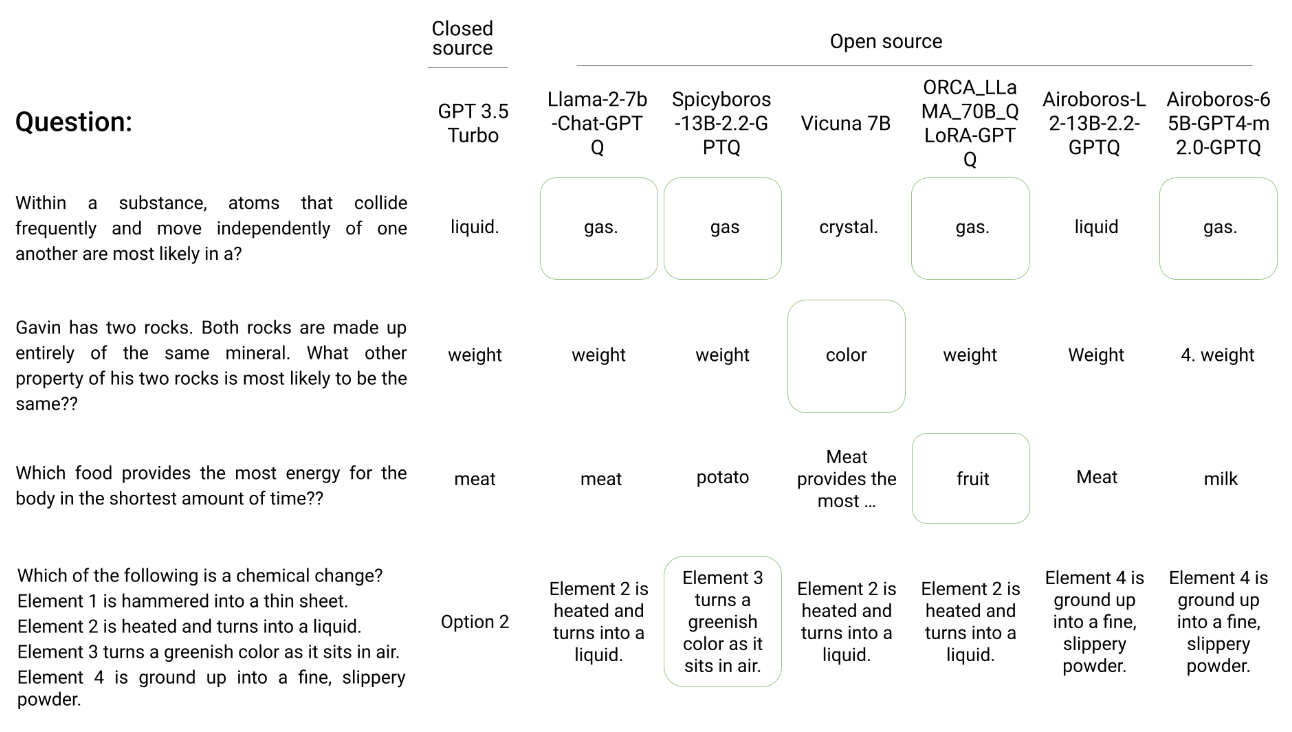
Herd: Using multiple, smaller LLMs to match the performances of proprietary, large LLMs via an intelligent composer
2023, arXiv preprint
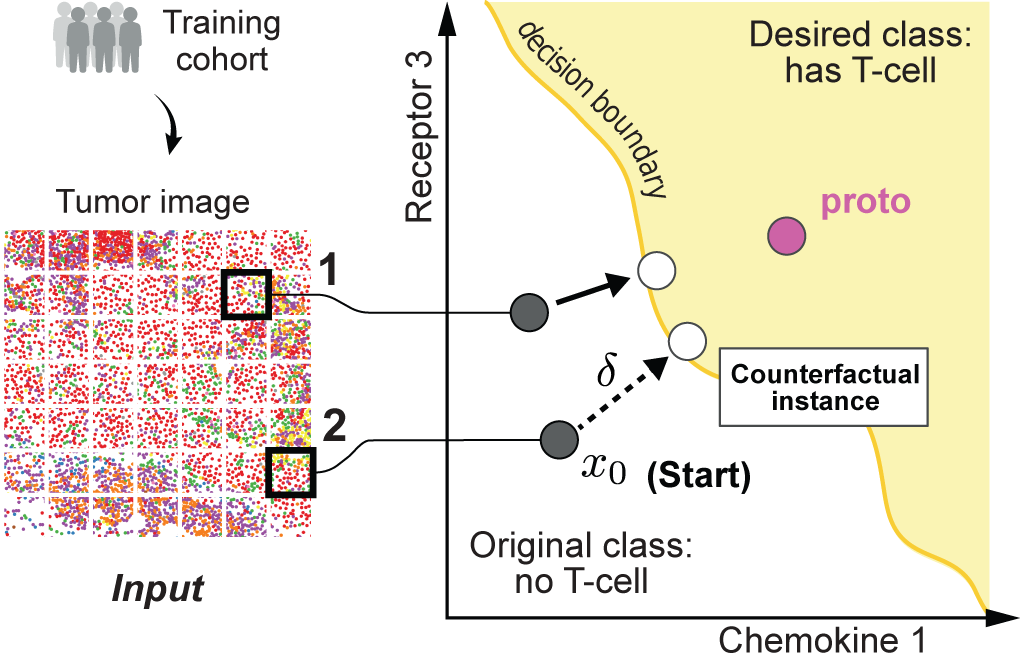
Generating counterfactual explanations of tumor spatial proteomes to discover therapeutic strategies for enhancing immune infiltration
2023, bioRxiv preprint
Tryage: Real-time, intelligent routing of user prompts to large language models
2023, arXiv preprint
Unraveling cell differentiation mechanisms through topological exploration of single-cell developmental trajectories
2023, bioRxiv preprint
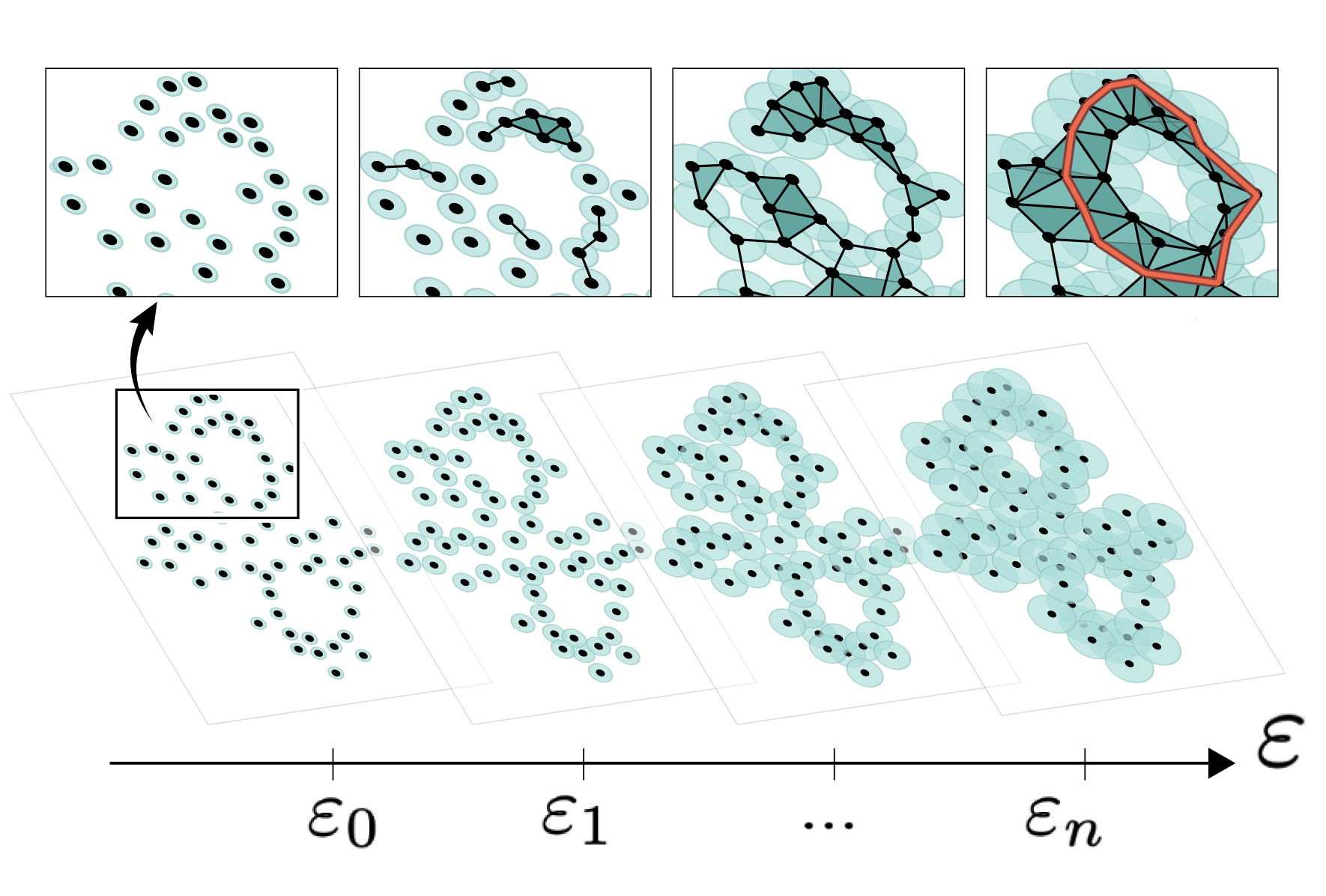
Dynamic Flow Control Through Active Matter Programming Language
2023, arXiv preprint
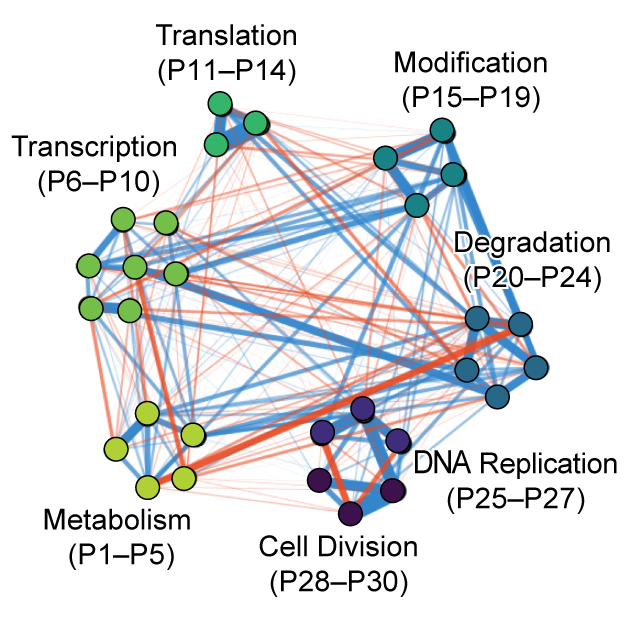
D-SPIN constructs gene regulatory network models from multiplexed scRNA-seq data revealing organizing principles of cellular perturbation response
2023, bioRxiv preprint
Connectedness of loss landscapes via the lens of Morse theory
2023, Proceedings of the 1st NeurIPS Workshop on Symmetry and Geometry in Neural Representations
Motor processivity and speed determine structure and dynamics of microtubule-motor assemblies
Rachel A Banks, Vahe Galstyan, Heun Jin Lee, Soichi Hirokawa, Athena Ierokomos, Tyler D Ross, Zev Bryant, Matt Thomson, Rob Phillips
2023, eLife 12:e79402
Spin glasses, error correcting codes, and synchronization of human stem cell organoids
2023, Cell (Preview Article)
2022
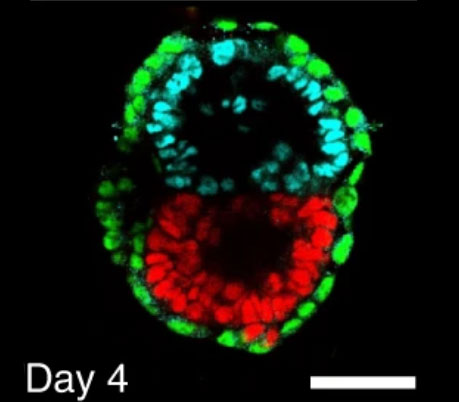
Stem cell-derived synthetic embryos self-assemble by exploiting cadherin codes and cortical tension
2022, Nature Cell Biology
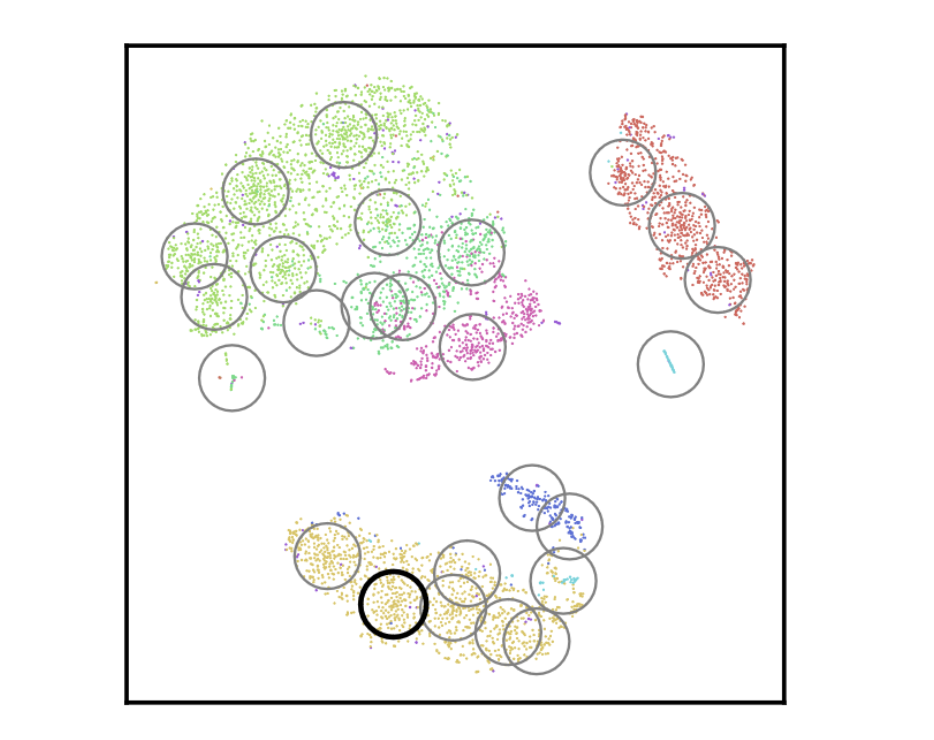
CloudPred: Predicting Patient Phenotypes From Single-cell RNA-seq
2022, Pacific Symposium on Biocomputing
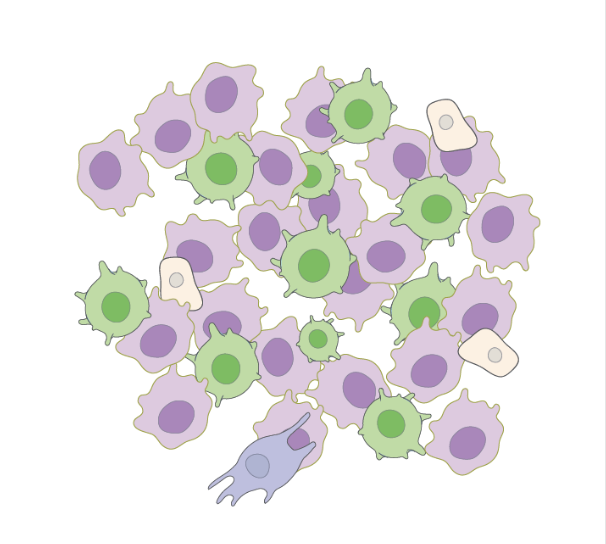
Generating counterfactual explanations of tumor spatial proteomes to discover effective, combinatorial therapies that enhance cancer immunotherapy
2022, NeurIPS AI for Science
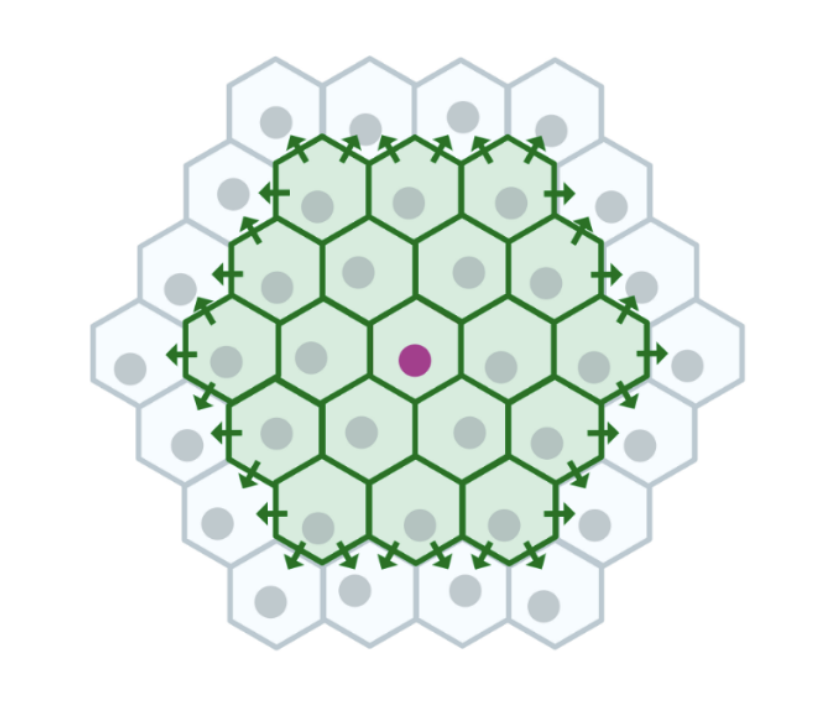
Neural networks learn an environment's geometry in latent space by performing predictive coding on visual scenes
2022, NeurIPS Information-theoretic Principles in Cognitive Science
Control of spatio-temporal patterning via cell density in a multicellular synthetic gene circuit
2022, bioRxiv
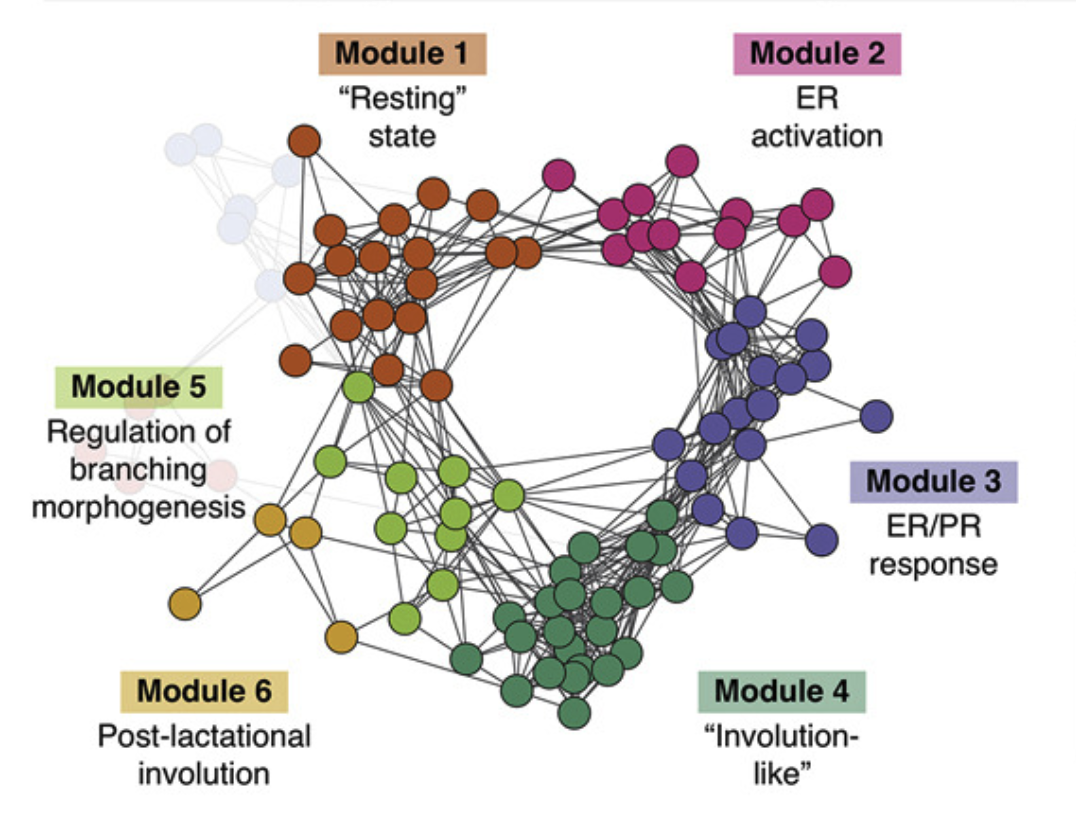
Mapping hormone-regulated cell-cell interaction networks in the human breast at single-cell resolution
2022, Cell Systems
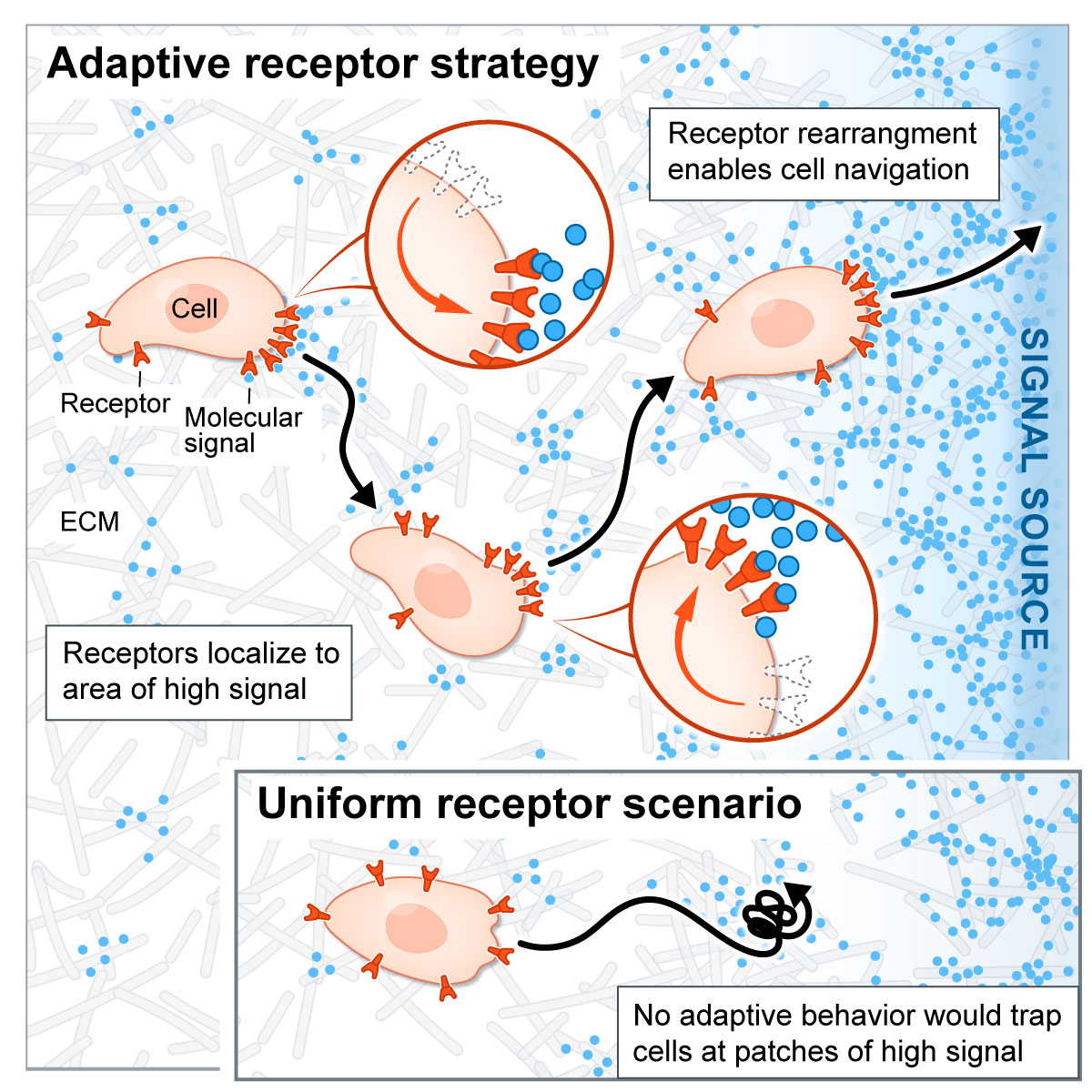
Localization of signaling receptors maximizes cellular information acquisition in spatially structured natural environments
2022, Cell Systems
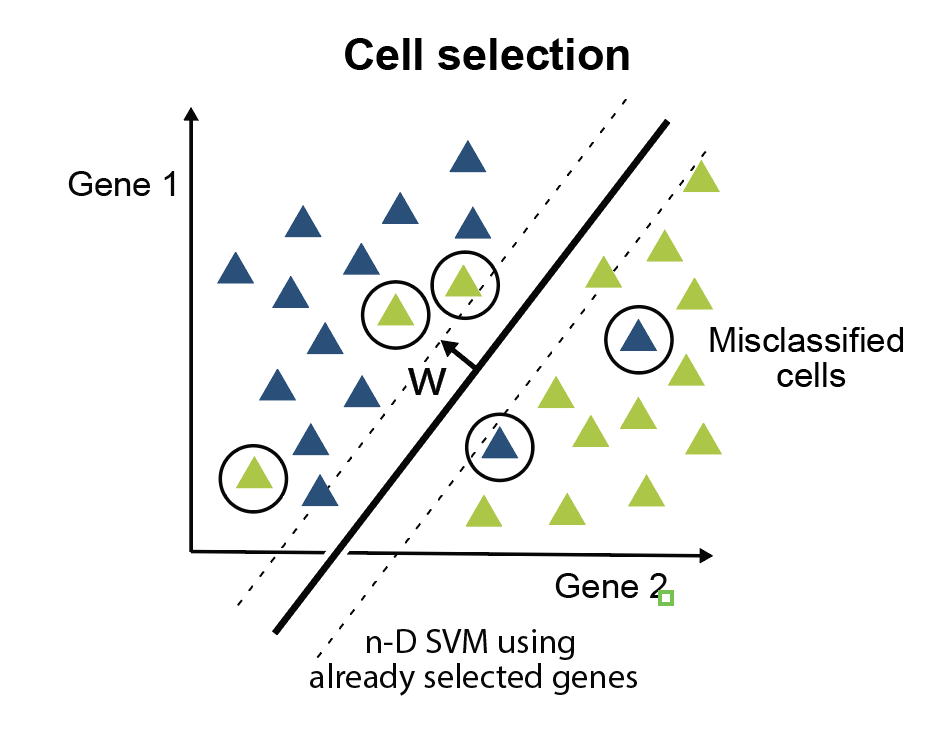
Minimal gene set discovery in single-cell mRNA-seq datasets with ActiveSVM
2022, Nature Computational Science
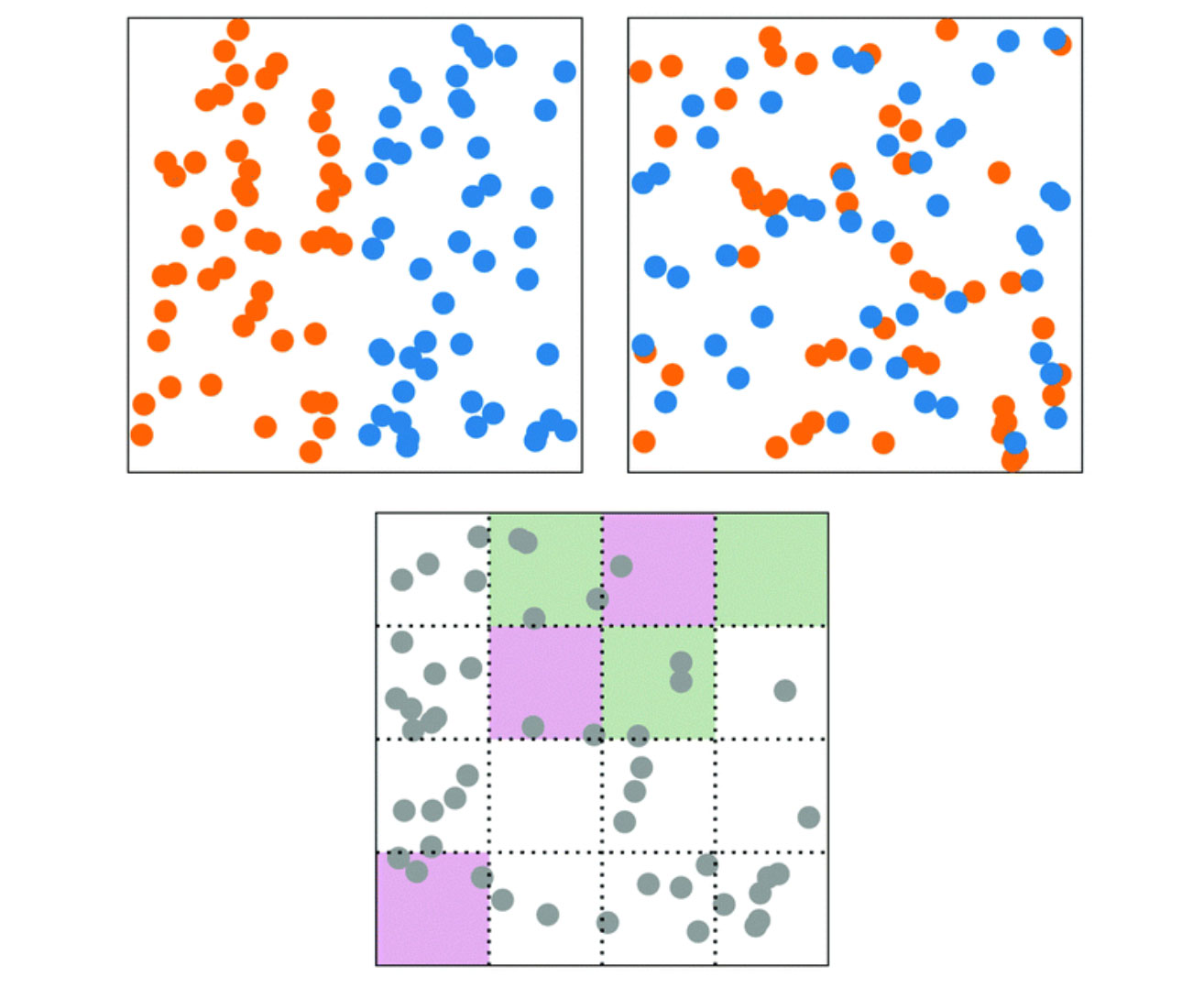
Reinforcement learning reveals fundamental limits on the mixing of active particles
2022, Soft Matter
A DNA repair pathway can regulate transcriptional noise to promote cell fate transitions
2022, Science
2021
Persistent fluid flows defined by active matter boundaries
Deep parallel characterization of AAV tropism and AAV-mediated transcriptional changes via single-cell RNA sequencing
Reconstructing aspects of human embryogenesis with pluripotent stem cells
Linear Transformations in Autoencoder Latent Space Predict Time Translations in Active Matter System
Enrique Amaya, Shahriar Shadkhoo, Dominik Schildknecht & Matt Thomson
NeurIPS AI for Science, 2021
Deep parallel characterization of AAV tropism and AAV-mediated transcriptional changes via single-cell RNA sequencing
Phenomenological model of motility by spatiotemporal modulation of active interactions
Dominik Schildknecht, Matt Thomson
New Journal of Physics, 2021
Active feature selection discovers minimal gene-sets for classifying cell-types and disease states in single-cell mRNA-seq data
Xiaoqiao Chen, Sisi Chen, and Matt Thomson
arXiv preprint, 2021
Solving hybrid machine learning tasks by traversing weight space geodesics
Reinforcement Learning reveals fundamental limits on the mixing of active particles
Dominik Schildknecht, Anastasia N. Popova, Jack Stellwagen, and Matt Thomson
arXiv preprint, 2021
2020
Sparsifying Networks by Traversing Geodesics
Developmental clock and mechanism of de novo polarization of the mouse embryo
Single cell profiling of capillary blood enables out of clinic human immunity studies
Tatyana Dobreva, David Brown, Jong Hwee Park, and Matt Thomson
Scientific Reports, 2020
Dissecting heterogeneous cell populations across drug and disease conditions with PopAlign
Programming Boundary Deformation Patterns in Active Networks
Persistent fluid flows defined by active matter boundaries
Self-organization of multi-layer spiking neural networks
Guruprasad Raghavan, Cong Lin, Matt Thomson
arXiv preprint, 2020
Geometric algorithms for predicting resilience and recovering damage in neural networks
Guruprasad Raghavan, Jiayi Li, Matt Thomson
arXiv preprint, 2020
Designing signaling environments to steer transcriptional diversity in neural progenitor cell populations
Jong H. Park, Tiffany Tsou, Paul Rivaud, Matt Thomson and Sisi Chen
bioRxiv, 2020
2019 – 2008
Highly Multiplexed Single-Cell RNA-seq for Defining Cell Population and Transcriptional Spaces
Controlling Organization and Forces in Active Matter Through Optically-Defined Boundaries
Active Learning of Spin Network Models
Jialong Jiang, David A. Sivak and Matt Thomson
arXiv preprint, 2019
Neural networks grown and self-organized by noise
Diffusion as a Ruler: Modeling Kinesin Diffusion as a Lenth Sensor for Intraflagellar Transport
Adult Neurogenesis Is Sustained by Symmetric Self-Renewal and Differentiation
Diffusion as a Ruler: Modeling Kinesin Diffusion as a Length Sensor for Intraflagellar Transport
Diffusion as a ruler: Modeling kinesin diffusion as a length sensor for intraflagellar transport
Transient Thresholding: A Mechanism Enabling Noncooperative Transcriptional Circuitry to Form a Switch
SOX2O-GlcNAcylation alters its protein-protein interactions and genomic occupancy to modulate gene expression in pluripotent cells.
Low Dimensionality in Gene Expression Data Enables the Accurate Extraction of Transcriptional Programs from Shallow Sequencing
Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors
Signaling Boundary Conditions Drive Self-Organization of Human “Gastruloids”.
Matthew Thomson
Developmental Cell, 39 (3). pp. 279-280. ISSN 1534-5807, 2016
