Links & Resources
Herd: The power of open-source language models en masse
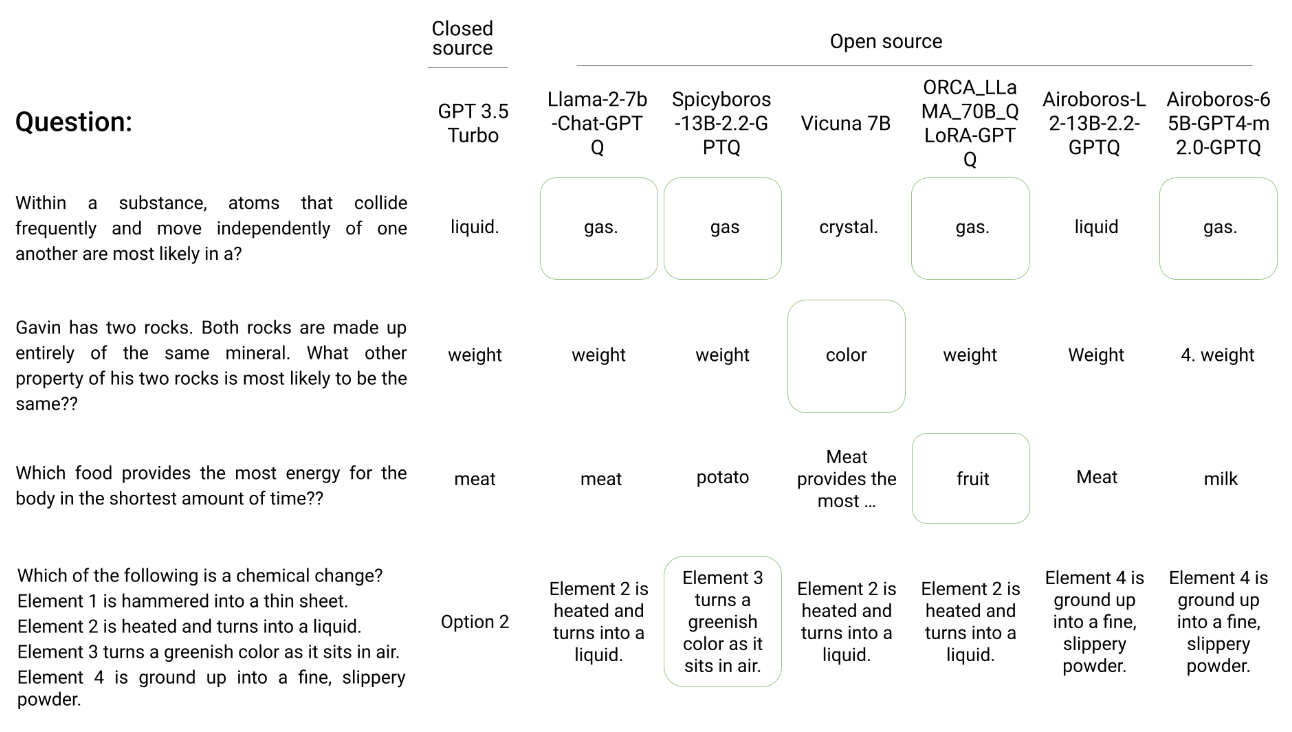
Could a herd of open source language models rival the performance of proprietary LLMs? Indeed, queries and responses using our Herd model router match the accuracy of ChatGPT, despite being composed of models that are effectively 2.5x smaller. Herd operates effectively at a fraction of the compute cost and zero query cost. Further, when proprietary models cannot answer a query, a herd of open source models are able to cover a significant portion of the deficit. This system offers a new model paradigm to compete against closed source models by leveraging widely available open source technology.
ActiveSVM: An active learning approach for compact gene set discovery
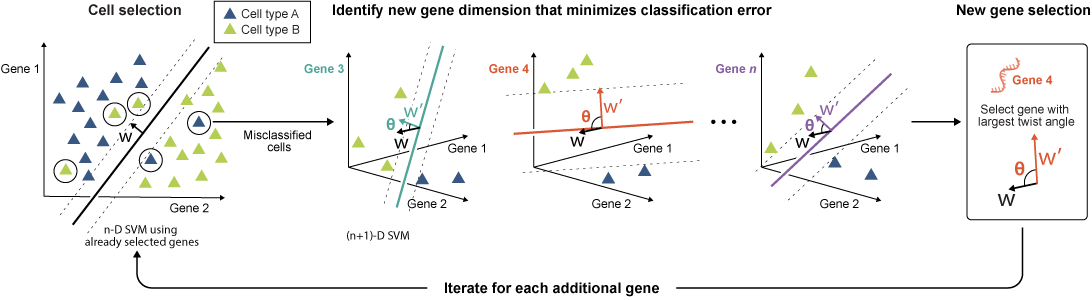
We introduce an active learning method that identifies minimal yet informative gene sets for classifying cell types, states, and perturbations in single-cell data using fewer genes. Our active feature selection employs an active support vector machine (ActiveSVM) classifier to generate compact gene sets from single-cell data. ActiveSVM identifies gene sets enabling ~90% cell-type classification accuracy across datasets. Discovering small but highly informative gene sets may reduce necessary measurements for single-cell mRNA-seq applications in clinical tests, therapeutic discovery, and genetic screens. Based on ActiveSVM, we present ActiveCell Inference, an end-to-end pipeline using ordered gene sets for fast and low-cost spatial genomics measurements by identifying well-classified cells requiring no further probing, reducing measurement costs 10 to 100-fold.
TRILL: Creative protein design in one platform
Many fields have rapidly adopted deep-learning models, partly due to the deluge of data humanity has amassed. In particular, the petabases of biological sequencing data enable the unsupervised training of protein language models that learn the “language of life.” However, due to their prohibitive size and complexity, contemporary deep-learning models are often unwieldy, especially for scientists with limited machine-learning backgrounds.
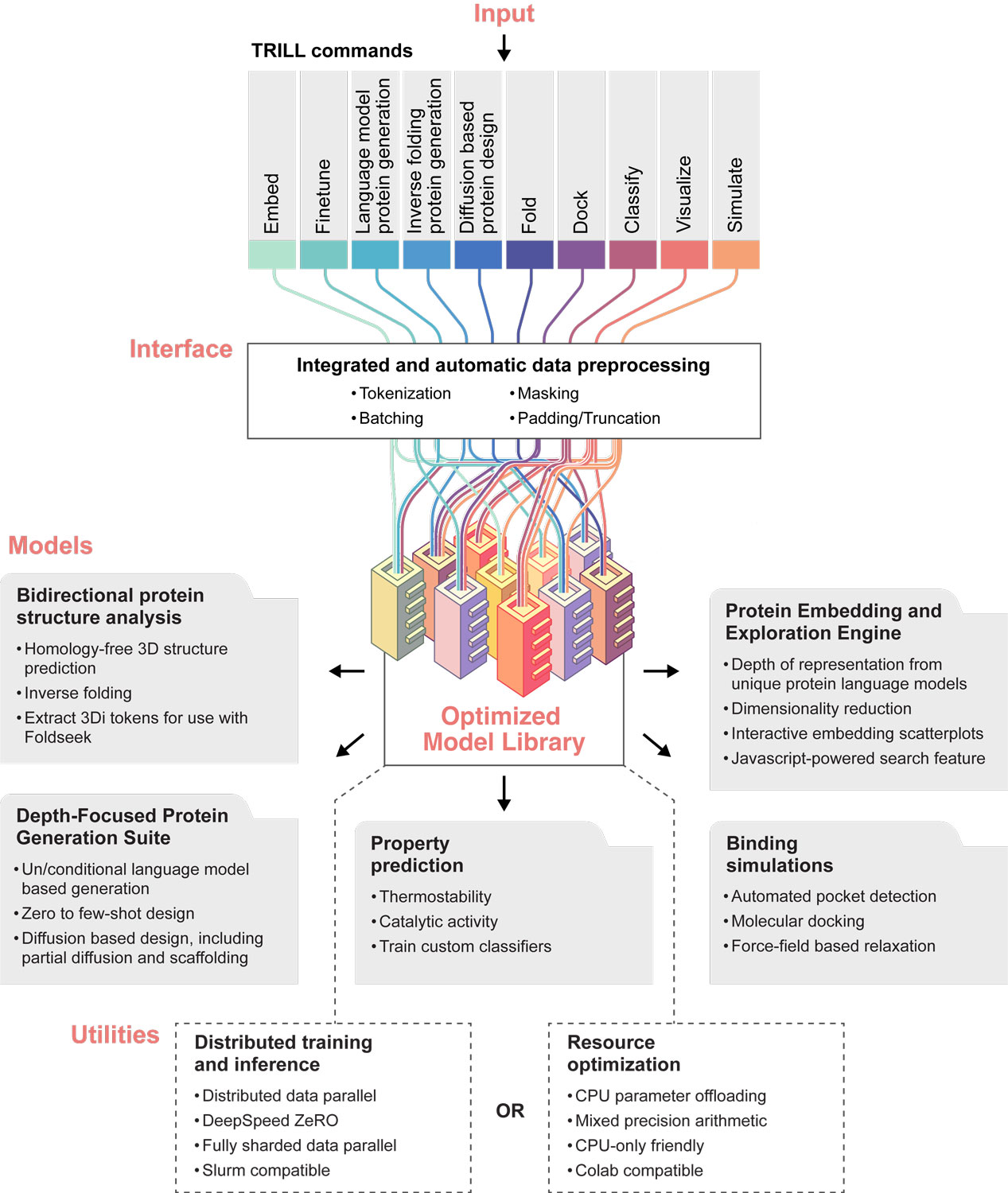
TRILL (TRaining and Inference using the Language of Life) is a platform for creative protein design and discovery. Leveraging several state-of-the-art models such as ESM-2, DiffDock, and RFDiffusion, TRILL allows researchers to generate novel proteins, predict 3-D structures, extract high-dimensional representations of proteins, functionally classify proteins, and more.
What sets TRILL apart is its ability to enable complex pipelines by chaining together models and effectively merging the capabilities of different models to achieve a sum greater than its individual parts. Whether using Google Colab with one GPU or a supercomputer with hundreds, TRILL allows scientists to effectively utilize models with millions to billions of parameters by using optimized training strategies such as ZeRO-Offload and distributed data-parallel. Therefore, TRILL not only bridges the gap between complex deep-learning models and their practical application in the field of biology, but also simplifies the orchestration of these models into comprehensive workflows, democratizing access to powerful methods.
Read the publication ⭢
Watch Thomson Lab videos on YouTube